Our humoral immune response is mediated by antibodies and triggered when antigens are detected through the binding of their B-cell epitopes to surface receptors of B lymphocytes. Most B-cell epitopes are conformational, formed by non-contiguous amino acid residues brought together by complex folding of the antigen. However, a small fraction are linear epitopes that are simpler, linear stretches of amino acid residues. Linear B-cell epitopes are ideal candidates in aiding vaccine formulation since they can trigger an immune response without proper folding and complex 3D conformation requirements.
Synthetic peptides containing B-cell epitopes administered to animals have shown to stimulate their immune system to secrete antibodies that recognize antigens from which these epitopes are derived. Therefore, the use of multiple synthetic peptides in formulating a vaccine is a highly promising approach in the development of subunit vaccines. These peptide-based vaccines have many advantages, including being both safer and more cost effective than attenuated and inactivated vaccines. Several peptide-based vaccines are currently under development for the prevention of infectious diseases such as malaria, influenza, and HIV. However, to identify B-cell epitope candidates rapidly and efficiently remains a formidable challenge.
Current identification strategies tend to involve the experimental detection of epitopes on a single protein, which are low throughput (one protein at a time) and require initial knowledge of the antigen and its sequence. High throughput strategies are available, but they require extensive use of synthetic biology and other time-consuming methodologies, e.g., library construction, peptide/protein array preparation and expression of proteins using non-native expression systems.
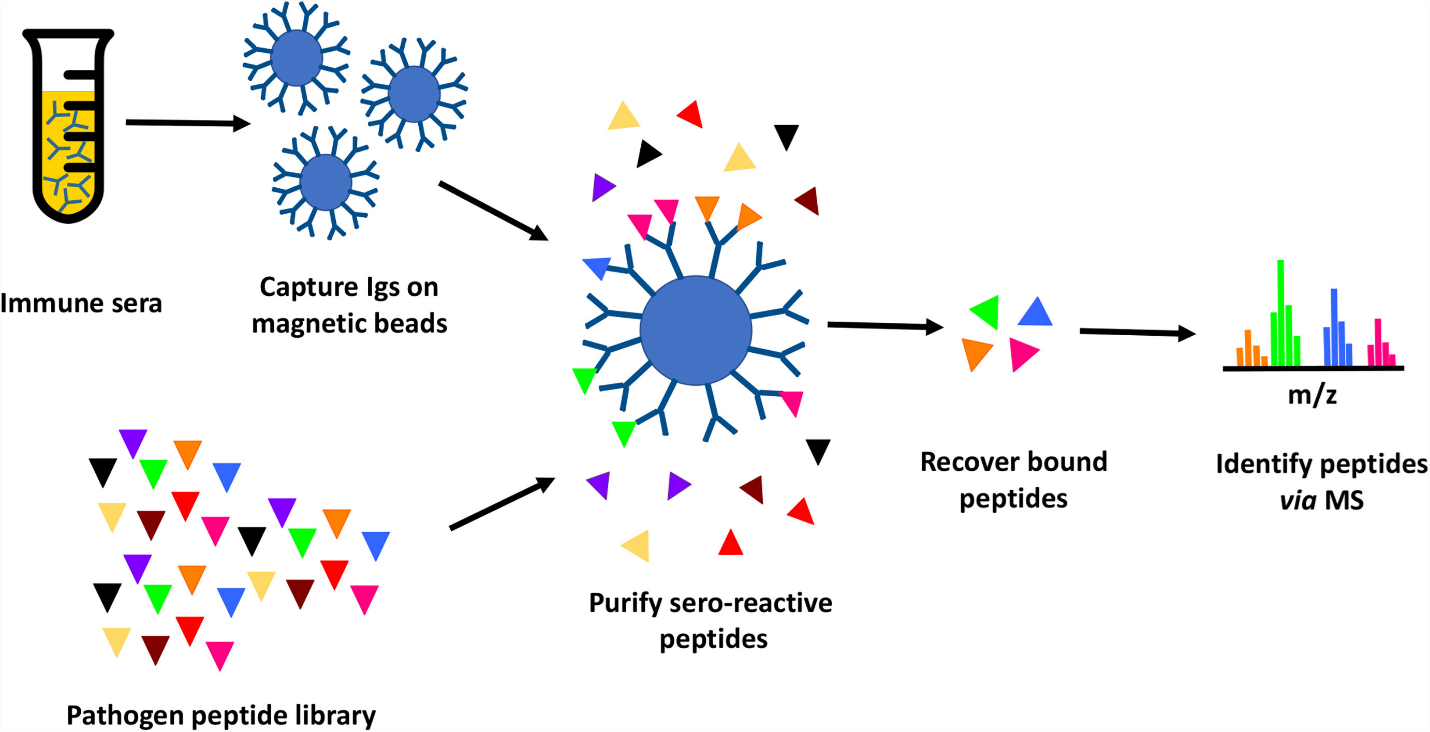
A more practical approach to characterize B-cell epitopes for peptide-based vaccine formulation would have to be simple, efficient, cost-effective and high throughput. Researchers at LLNL have developed a shotgun immunoproteomic approach that can fulfil these requirements while also be broadly applicable to the identification of linear B-cell epitopes from any pathogen.
LLNL’s high throughput method involves proteome-wide screening for linear B-cell epitopes using native proteomes isolated from a pathogen of interest and convalescent sera from immunized animals. LLNL researchers have applied their newly developed generalizable screening method to the identification of pathogenic bacteria by screening linear B-cell epitopes in the proteome of Francisella tularensis as well as Burkholderia pseudomallei; both are considered Tier 1 Select Agents by the Centers for Disease Control and Prevention. The results from these two test cases showed the screening method was successful in identifying peptides that map to known antigens of these infectious organisms.
LLNL’s novel immunoproteome screening method used in the test cases can easily be adapted to detecting peptide targets relevant to the immune systems of other mammalian species, including humans (depending upon the availability of convalescent sera from patients), and could aid in accelerating the discovery of B-cell epitopes and development of effective vaccines and therapeutics for a variety of diseases.
D'haeseleer P, Collette NM, Lao V, Segelke BW, Branda SS, Franco M. Shotgun Immunoproteomic Approach for the Discovery of Linear B-Cell Epitopes in Biothreat Agents Francisella tularensis and Burkholderia pseudomallei. Front Immunol. 2021 Sep 29. (https://doi.org/10.3389/fimmu.2021.716676)
LLNL’s high throughput method for epitope discovery has many advantages:
- Is a high throughput method.
- Prior knowledge of antigenicity and structural information not required.
- Does not involve laborious experimental techniques as compared to technologies currently available.
- Designed to provide not only the protein antigen identification, but (more importantly) also delineate the antigenic regions of the identified antigens (epitopes).
- Facilitate development of peptide-based vaccines, which are safer and more cost effective than attenuated and inactivated vaccines.
Peptide-based vaccine and therapeutic development to counter biothreat agents, emerging pathogens and other pathogens of interest
Current stage of technology development: TRL-2
LLNL has patent protection on this invention.
U.S. Patent No. 11492380 PepCon proteomics standards and methods of use published 11/8/2022
US Patent Application Publication No. 20230192779 PEPCON PROTEOMICS STANDARDS AND METHODS OF USE published 6/22/2023