There are trillions of unique microorganisms on this planet and identifying and analyzing specific viruses, pathogens, or other microorganisms in clinical or environmental samples can be challenging. Culturing viruses and certain other microorganisms for differentiation analysis is difficult. Few, if any, repeated sequences between microorganism genomes indicates fickle lineage between species and confounds the limited information available on, say, viral and other pathogen backgrounds. Metagenomics has been beneficial in providing metagenomic libraries, however, many metagenomic sequences have no homologues present in their databases, a key identifying factor for organisms. Improving the ability to identify and differentiate between organism families plays a key part in epidemic preparation and response.
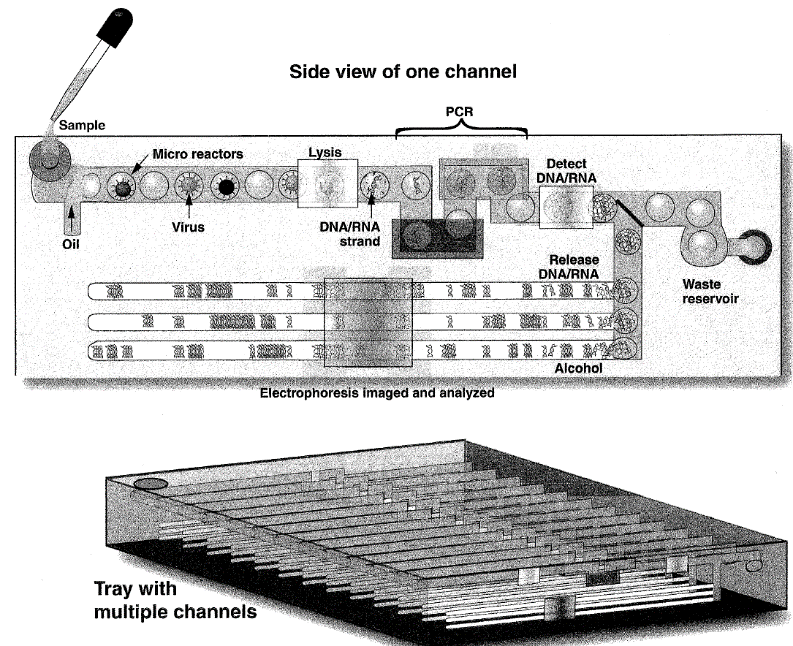
LLNL researchers have invented a system for identifying all known and unknown pathogenic or non-pathogenic organisms in a sample. This invention takes a complex sample and generates droplets from it. The droplets consist of sub-nanoliter volume reactors which contain the organism sized particles. A lysis device lyses the organisms and releases the nucleic acids. An amplifier then magnifies the quantity of available nucleic acids. Then, a fractionator liberates the nucleic acids from the droplets. Finally, a parallel analyzer identifies all the known and unknown pathogen or non-pathogenic organisms in the complex sample. This device functions with DNA or RNA samples.
- Speedy and accurate detection and identification of organisms
- Parallel analyzer is compact for clinical, field, or research purposes
- Device can function with small sample sizes
- Identification and detection of bio-threat agents which contain nucleic acid signatures such as spores, bacteria, and viruses
- Identification and monitoring of outbreaks of infectious diseases including emerging or unknown pathogens
- Detection of host or microbial and viral DNA or RNA in biological clinical samples
LLNL currently holds patent 8,338,166 "Sorting, amplification, detection, and identification of nucleic acid subsequences in a complex mixture" for this technology (LLNL internal # IL-11652).