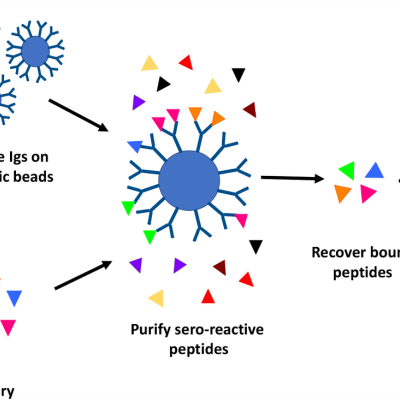
LLNL’s high throughput method involves proteome-wide screening for linear B-cell epitopes using native proteomes isolated from a pathogen of interest and convalescent sera from immunized animals. LLNL researchers have applied their newly developed generalizable screening method to the identification of pathogenic bacteria by screening linear B-cell epitopes in the proteome of Francisella…
Keywords
- Show all (81)
- Instrumentation (38)
- Diagnostics (13)
- Data Science (5)
- Therapeutics (5)
- Cybersecurity (4)
- Brain Computer Interface (BCI) (3)
- Computing (2)
- Imaging Systems (2)
- Simulation (2)
- Analysis (1)
- Polymer Electrodes (1)
- Quantum Science (1)
- Rare Earth Elements (REEs) (1)
- (-) Vaccines (2)
- (-) Information Technology (1)
Technology Portfolios

LLNL has developed a new active memory data reorganization engine. In the simplest case, data can be reorganized within the memory system to present a new view of the data. The new view may be a subset or a rearrangement of the original data. As an example, an array of structures might be more efficiently accessed by a CPU as a structure of arrays. Active memory can assemble an alternative…
LLNL has developed a novel process of production, isolation, characterization, and functional re-constitution of membrane-associated proteins in a single step. In addition, LLNL has developed a colorimetric assay that indicates production, correct folding, and incorporation of bR into soluble nanolipoprotein particles (NLPs).
LLNL has developed an approach, for formation of NLP/…
